| 类型 | 说明 |
|---|---|
| 论文信息 | U-Net: Convolutional Networks for Biomedical Image Segmentation Olaf Ronneberger, Philipp Fischer, and Thomas Brox |
| 会议期刊 | MICCAI 2015 |
| 作者所在机构 | 德国弗莱堡大学 |
| 解决的问题 | 生物医学分割任务 |
| 模型名 | U-Net |
| 模型结构 | 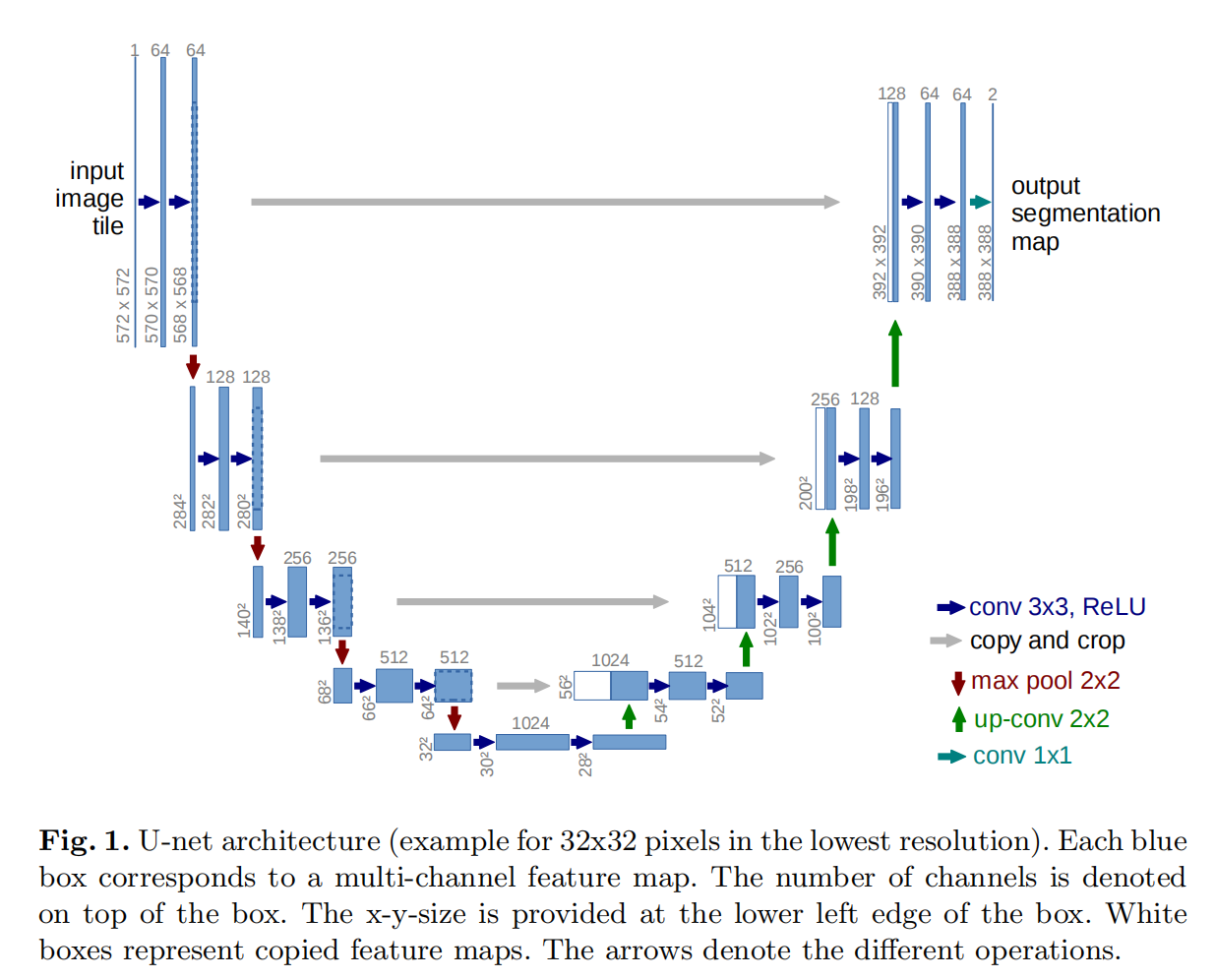 |
| 模型特点 | 1.U字型结构,上采样需要用到前四个Block生成的特征图 2.注意到卷积层No-padding,所以下采样Block产生的特征图大小与上采样后的特征图大小不一致,需要裁减 |
| 数据集 | ISBI 2012 挑战赛数据集(注册页面404,无法获取) |
| 数据增强 | 使用3×3的随机替换的平滑变换 每个像素的替换使用双三次差值进行计算 |
| 相关参数 | SGD优化算法,momentum=0.99,batchsize=1,框架caffe 初始化:标准差为$\sqrt{\frac{2}{N}}$的高斯分布初始化权重, $N$为输入节点个数 |
| Softmax | $$p_k\left(x\right) = \frac{e^{a_{k}\left(x\right)}}{\sum_{k^{\prime} = 1}^{K} e^{a_{k}^{\prime}\left(x\right)}}$$ 1.$x\in\Omega, \Omega\subset Z^{2}$:x为每个像素 2.$a_{k}\left(x\right)$:每个像素点x在对应特征通道k的得分 3.$K$为特征通道数 $p_{k}为对于特征通道即类K的预测分类结果$ |
| 交叉熵函数 | $$E = \sum_{x\in\Omega}w\left(x\right)log\left(p_{\ell\left(x\right)}\left(x\right)\right)$$ 1.$\ell: \Omega \rightarrow \lbrace 1,…,K \rbrace$:每个像素的真是标签 2.$\omega:\Omega\rightarrow R$:权重图 |
| 权重图 | $$w\left(x\right) = w_{c}\left(x\right)+w_0\cdot exp\left(-\frac{\left(d_{1}\left(x\right)+d_{2}\left(x\right)^{2}\right)}{2\sigma^{2}}\right)$$ 1.$w_{c}$:平衡每个类频率的权重图 2.$d_{1}:\Omega\rightarrow R$:距离最近细胞边界的距离 3.$d_{2}:\Omega\rightarrow R$:距离次近细胞边界的距离 4.$w_{0}=10, \sigma \approx 5$ |
| 实验 | — |
| 1 | 电子显微镜图像的神经元分割 |
| 2 | 胶质母细胞瘤与海拉细胞分割实验 |
| 结论 | 1.实现了不同生物医学分割应用上的非常好的性能 2.仅仅需要少量标注图像以及可接受的合理训练时间。 |
| 思考 | 1.卷积层No-padding导致输出图像与原图不一致,故在其他应用是可以考虑加上padding=1以获得与输入相同大小图像 2.数据集全来自ISBI,需要进一步在不同机构采集的图像上实验 3.可以考虑将此模型应用于其他任务,例如分类,和目标检测等的实验 |
| Achitecture | Specification |
|---|---|
| Model Component | |
| BaseConvBlock | 1.Convolutional Layer:channels=output_channels, kernel_size=3, padding=0, strides=1 2.ReLu Activation Layer 3.Convolutional Layer: channels=output_channels, kernel_size=3, padding=0, strides=1 4.ReLu Activation Layer(Note: no-padding) |
| DownSampleBlock | 1.Max Pooling Layer:pool_size=2, strides=2 2.BaseConvBlock(output_channels) |
| UpSampleBlock | 1.Deconvolutional Layer(or Upsampling Layer):channels=output_channels, kernel_size=2, strides=2 2.BaseConvBlock(output_channels) |
| Model | |
| Contracting Path | |
| InputBlock | BaseConvBlock(output_channels),Output: Feature Map X1 |
| 1-th DownSampleBlock | output_channels=128, Output: Feature Map X2 |
| 2-th DownSampleBlock | output_channels=256, Output: Feature Map X3 |
| 3-th DownSampleBlock | output_channels=512,Output: Feature Map X4 |
| 4-th DownSampleBlock | output_channels=1024,Output: Feature Map X5 |
| Expanding Path | |
| 1-th UpSampleBlock | output_channels=512 A new feature map is obtained by connecting Feature Map from deconvolutional layer and Feature Map X4(need to crop) according to the channel dimension, which is used as the input of its BaseConvBlock |
| 2-th UpSampleBlock | output_channels=256 A new feature map is obtained by connecting Feature Map from deconvolutional layer and Feature Map X3(need to crop) according to the channel dimension, which is used as the input of the BaseConvBlock |
| 3-th UpSampleBlock | output_channels=128 A new feature map is obtained by connecting Feature Map from deconvolutional layer and Feature Map X2(need to crop) according to the channel dimension, which is used as the input of the BaseConvBlock |
| 4-th UpSampleBlock | output_channels=64 A new feature map is obtained by connecting Feature Map from deconvolutional layer and Feature Map X1(need to crop) according to the channel dimension, which is used as the input of the BaseConvBlock |
| OutputBlock | 1×1 Convolutional Layer:output_channels=num_classes |
模型实现
1 | # U-Net Model |
模型各层输出形状(B×C×H×W)
1 | Contracting Path: |
1 | Contracting Path: |
